The evolution of fungal secondary metabolism
COLLABORATORS: The Keller lab (UW Madison), The Branco Lab (UCO Denver)
Soil fungi are prolific producers of bioactive metabolites, responsible for the production of some of our most important pharmaceuticals and therapeutics. Antagonism and stress within rhizosphere communities drives the evolution of this novel biochemical diversity. This project uses genomics, bioassays, and metabolomics to characterize the functional and ecological roles of fungal bioactive metabolites, and the evolutionary history and genomic innovations responsible for the evolution of fungal secondary metabolism.
The population ecology of fungal pathogens
COLLABORATORS: The Stajich lab (UCR), The Cramer Lab (dartmouth)
Aspergillus fumigatus is a ubiquitous fungal saprophyte, and one of the most important causative agents of human fungal infections across the world. This project uses a combination of reference-based and reference-free strategies including pan-, population-, and phylo-genomics, to characterize population structure and genomic drivers of population differentiation, recombination frequency, and capacity for in-host evolution in A. fumigatus.
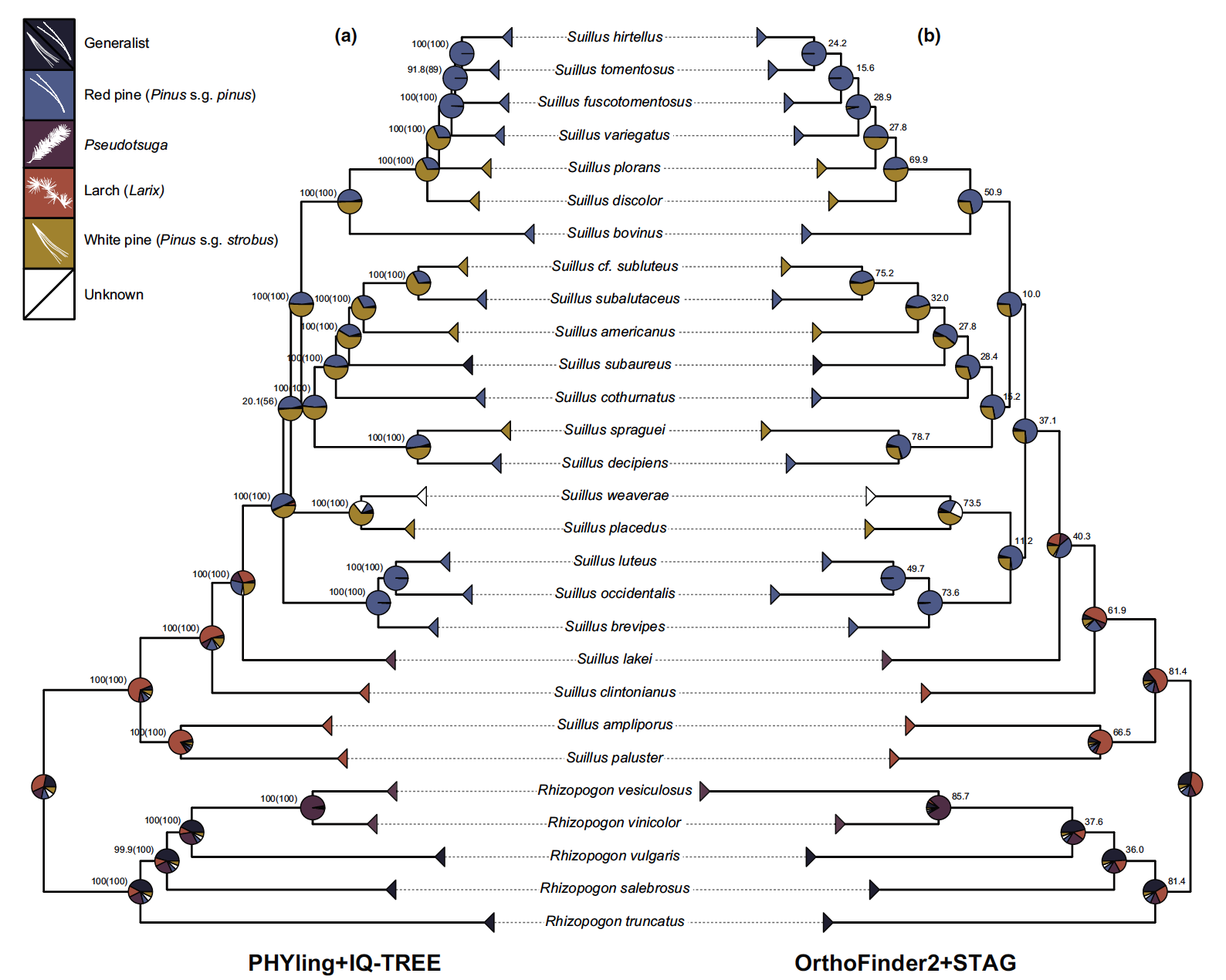
Host-specificity & host-switching
COLLABORATORS: The Suillus Consortium, THE JGI
A large number of fungal species are incapable of surviving on their own. These obligate symbionts depend on other species for their survival, and represent a range of ecological guilds- from deadly pathogens, to vital mutualists. In a series of projects spanning the fungal phylogeny, we are using comparative genomics and high throughput multi-omics to investigate the evolution and genetic mechanisms structuring host-range, host-specificity and host-switching in fungal symbionts.
Context dependent guild occupation
COLLABORATORS: The kistler lab (USDA-ARS)
Fusarium graminearum contaminates wheat and barley crops by producing a family of mycotoxins known as trichothecenes. Fusarium spp. producing B-type trichothecenes are thought to have evolved in North America in the middle Pliocene, approximately 3.5 MYA. As such, grasses native to the United States grain belt have likely had a longer evolutionary history with F. graminearum than cultivated members of the Poaceae which were only introduced to North America by more recent human activity. Although Fusarium is easily isolated from the seed of native grass species, many native grasses fail to present the suite of symptoms typically associated with trichothecene accumulation in cultivated grasses. This project investigates the chemotypic distribution, evolutionary drivers and ecological consequences of Fusarium infection in native perennial prairie grasses growing in the United States' grain belt.